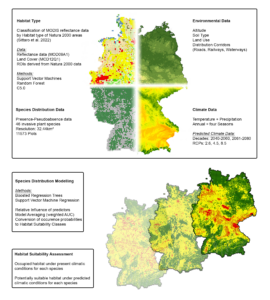
Determining the habitat suitability of invasive plant species
Our work to determine habitat suitability for invasive plant species in Germany is based on two fundamental procedures that we have explained in detail in two scientific publications - firstly, the remote sensing classification of habitat types based on satellite data, on the other hand the species distribution modelling, in which the occurrence of invasive plant species is related to the environmental conditions of the habitats they colonise. A brief description of the methods used can be found here - the two publications are linked at the end of the page.
Methodology
Publications
Which factors determine the invasion of plant species? Machine learning based habitat modelling integrating environmental factors and climate scenarios.
Sittaro F., Hutengs C., Vohland, M.:
International Journal of Applied Earth Observation and Geoinformation, 2023; 116. https://doi.org/10.1016/j.jag.2022.103158.